Lab Meeting on 5 September 2019
We were in a tiny meeting room at the Alan Turing Institute today because the Institute was hosting a Data Study Group. Many apologies to Konrad Adler-Wagstyl and Hannah Spitzer who had lots of questions from the many lab members who connected through Zoom rather than over fill the room with CO2! Thank you so much for coming and presenting your work on the amazing MELD project (see below for more details.)
Celebrations and cool things to share
This was Maxine’s last meeting. She’s finished her enrichment year at the Turing and is now writing up her thesis from her home in Oxford. Thank you so much for being part of the group, Maxine! We’ll miss you 💖
Kirstie had some really fun conversations at the INCF Assembly in Warsaw this week (including finding out that the team who collected the Study Forrest dataset have never analysed it, and never intended to!). She gave a talk on “Ten Simple Rules to Run an Open and Inclusive Project Online” (slides doi: 10.5281/zenodo.3383062, tweet thread) and made the picture below that she’s particularly proud of 😂. She was also really proud to be a member of the Turing Safe Haven project team who published their design paper on arXiv this week (arxiv: 1908.08737). Kirstie ran the first Turing Way online Collaboration Cafe this week and so appreciated Nadia, Jez and Malvika joining her to develop the Turing Way project (video: https://youtu.be/I0z7OEbBzes). Finally, she shared this blog post on survival skills for women (and other URMs in tech). A depressing but very useful read.
Sarah and Kirstie submitted a request for a portion of the Microsoft Azure credits donation to the Turing Institute to be used to build a BinderHub cluster at the Turing that will receive traffic from mybinder.org and be a part of the Binder Federation! 🎉🎉🎉
She also shared a website (HT Erik Sundell) that shows the cleanliness of a country’s energy generation and how the import/export of cleaner energy can effect the emission outputs of other countries that may use dirtier resources: https://www.electricitymap.org It’s an excellent piece of scientific communication: the colour scheme is easily understood, and leads to great discussions about which countries you could pair up to reduce the international level of emissions.
Elizabeth shared a very cool tool to “Manage Messes in Computational Notebooks”.
Ang promoted a project run by his PhD research institution (Institute of Automation, Chinese Academy of Sciences) to hire some excellent early career researchers in AI.
Patricia is settling into her new job and is celebrating the office having separate crisps package recycling bin 😋
Yini shared a paper showing that Wednesday is a lucky day to submit your paper to a journal!! doi: 10.1016/j.physa.2016.10.078, author accepted version.
Malvika celebrated being accepted to co-lead a Mozilla Open Leadership program in 2020 with Yo and Berenice Batut. They will be receiving training in the next months. She also shared some recent blog posts on community building:
- 5 tips to promote ‘water cooler effects’ at informal discussion sessions
- How to be a pessimistic organiser for successful events
Questions we’re thinking about
Maxine read this article asking “Do You Want to Be Known For Your Writing, or For Your Swift Email Responses?" and is wondering about the liberation of not having to aim for inbox zero all the time? Or is that just rude to colleagues and collaborators?
Patricia is trying to start some really organised habits for her new job and is looking for tips for collecting her notes together regularly.
Yini has been thinking about the work of the of the Brain Imaging Data Structure (BIDS, https://bids.neuroimaging.io) community in trying to unify the format of MRI data. Having data in one harmonised format makes it much easier to run analyses, but she’s wondering about the effects of a prevalent analysis pipeline that may have different preprocessing choices than other research groups would implement. These decisions will always influcent the results. How can we make sure that certain research questions are not ignored because the tools do not exist to ask them?
Ang has noticed that in many regions the individual measures for cortical thickness produced by Freesurfer correlate with the group patterns, but that isn’t always the case. It is probably a spatial bias in the accuracy of the recon-all algorithm. How can we (should we) detect subtle mis-alignments?
Malvika is still struggling to get some interviews transcribed. Does anyone have any suggestions to do this on and off line? (Some data can be used online, others can only be processed locally). Any motivation tips?
Konrad and Hannah presenting on the MELD project
We zoomed through all the updates above, and commented in our shared HackMD notepad before the meeting, because we wanted to have lots of time to learn more about the Multi-centre Epilepsy Lesion Detection (MELD) project
The goal of the project is to use machine learning to identify structural abnomalities on MRI for people with treatment resistant epilepsy. Sophie and Konrad analysed a smaller dataset a few years ago and managed to do very well on that cohort (doi: 10.1016/j.nicl.2016.12.030).
They now have 450 participants from 18 different research groups around the world.
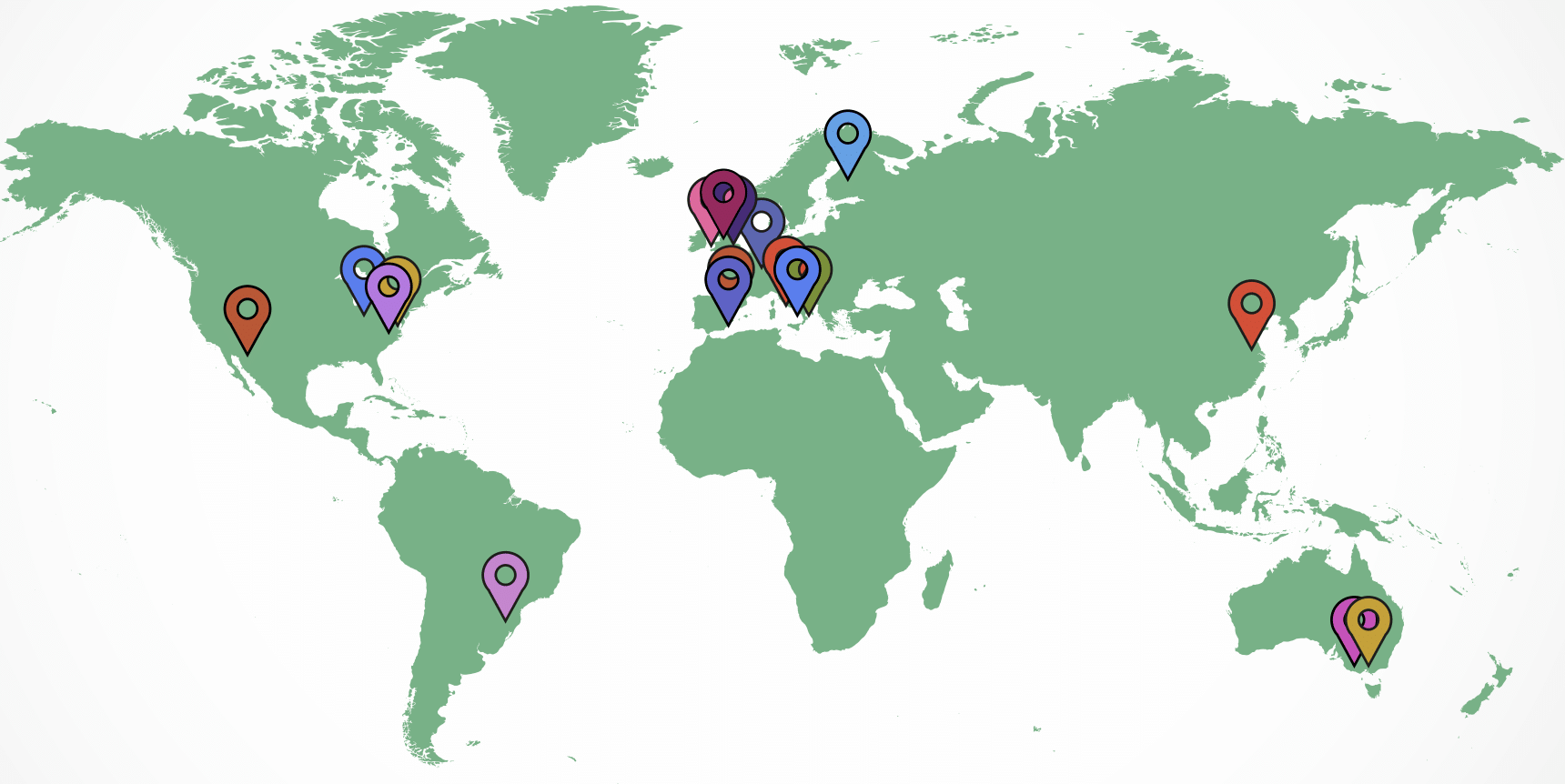
As data on patients with epilepsy is very sensitive - individual participants could be much more easily identified than people who do not have a neurological disorder - all the data processing happens locally at the sites where data was collected. Only fully anonymised data is transferred to the MELD team. Their collaborators around the world follow the detailed study protocols which are publicly available and licenced for re-used through protocols.io: https://www.protocols.io/researchers/meld-project/protocols.
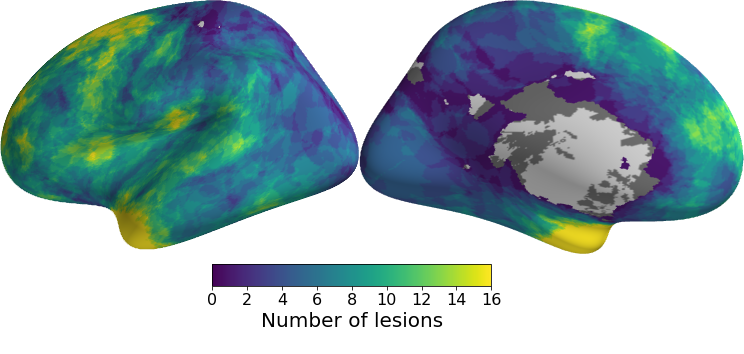
Cortical lesions are very small and it takes a traned expert to identify them. These labels are shipped with the anonymised data. You can see the location of the lesions across all members of the cohort in the picture below.
The lab meeting was rather like a second PhD viva for Konrad, with questions on MRI processing, quality control, data transfer and security, patient consent in a future national health service with interoperable data access, deep learning with mesh convolutions, normative modelling to avoid hand labelling of the data, and the benefits of multi-modal data 🙀🙀🙀.
Thank you so much Konrad and Hannah for coming along and sharing your work with us.
If you are interested in getting involved in this collaboration please do not hesitate to contact the MELD team at MELD.study@gmail.com. They are always looking for interested epilepsy centres as well as research scientists, developers and database managers.
